Relic DNA - Do Dead Cells Impact Microbiome Analysis?
- zackljones

- Dec 27, 2025
- 6 min read
A fairly common question I receive revolves around whether DNA sequencing detects organisms that are not active or dead. This is a very legitimate concern as DNA sequencing can detect DNA from dead organisms. This DNA can stay inside a dead cell for awhile and will eventually start floating around the environment if the cell or DNA is not consumed.

Names of DNA from dead cells
Before we dig into this topic I wanted to cover the different names you might here to refer to DNA from dead cells.
Extracellular DNA - This refers any DNA outside the cell, usually DNA from dead cells but can also include DNA purposely released by cells.
Environmental DNA (eDNA) - tends to refer to all DNA floating around the environment from all forms of life including dead cells from macroorgnaisms like fish and insects. This name is usually used when performing broad monitoring of all life like fish in river.
Relic DNA - the primary topic of this post and refers to the the pool of DNA from dead microorganisms that persists in the environment.
Advantage of Detecting DNA from Dead Cells
We typically use fact that DNA persists after a cell dies to our advantage during sample collection. The collection tubes Aggrego Data provide contain a preservative that both "fixes" the microbial community which is a nice way of saying it instantly kills everything or at least prevents them reproducing. This fixation prevents the microbial community from changing after sample collection and it also protects DNA from degrading even at room temperature. For the most part, this a big advantage over other microbial analysis techniques as it allows for reliable sample collection from all over the world.
The downside is that we might be picking up DNA from dead cells prior to collection which is what I want to explore today. How big of a problem is relic DNA and what are the implications for microbiome analysis?
Effect of Relic DNA on Microbiome Analysis in Soils
While DNA sequencing of microbial communities really started gained traction in 2009-2010, it wasn't until late 2016 that DNA from dead cells and it's impact on the results of DNA sequencing of microorganisms was investigated. This ground breaking study was led by Paul Carini out of the laboratory of Prof. Noah Fierer in Boulder, Colorado. The study looked at a variety of soil types and ask fairly simple questions - how much of the total microbial DNA was from dead cells and how did this relic DNA affect microbial diversity estimates?

As seen in the figure above, relic DNA on average account for ~40% of the total bacterial (a) and fungal (b) DNA recovered. However, this amount can vary considerably from one soil to the next with some soils having over 80% of the total DNA be from non-living organisms while other soils have close to 0% relic DNA.
These findings implicate that quantitative DNA based methods are likely to overestimate the total bacteria and fungi in many soils. This figure also suggests that relic DNA can increase diversity estimates (c and d) as the dead organisms might be not be the same species as the living organisms. However, this 2018 paper suggests that relic DNA does not affect diversity estimates and generally reflects the living microbial community. The authors also noted that relic DNA could create noise when looking for shifts in microbial communities over time or between treatments making these types of studies in soils even more challenging.
Soil Properties and Relic DNA Abundance
As the previous figure showed, there can be a huge variation in amount of relic DNA stored in soils. The primary soil characteristics that determine the abundance of relic DNA have to do with how well the DNA is protected. DNA itself is a pretty stable molecule, but will be readily consumed by microorganisms or broken down by enzymes.
Research has shown that the retention of relic DNA in soils is influenced by electrostatic interactions (positive and negative charges) between DNA and soil particles. This is complex topic that took me awhile to understand. It has to do with the amount of clays and organic matter in the soils as well as positively charged cations such as K+, Ca2+, and Na+. I'll try to break it down, but it's probably easiest to refer to the image below.
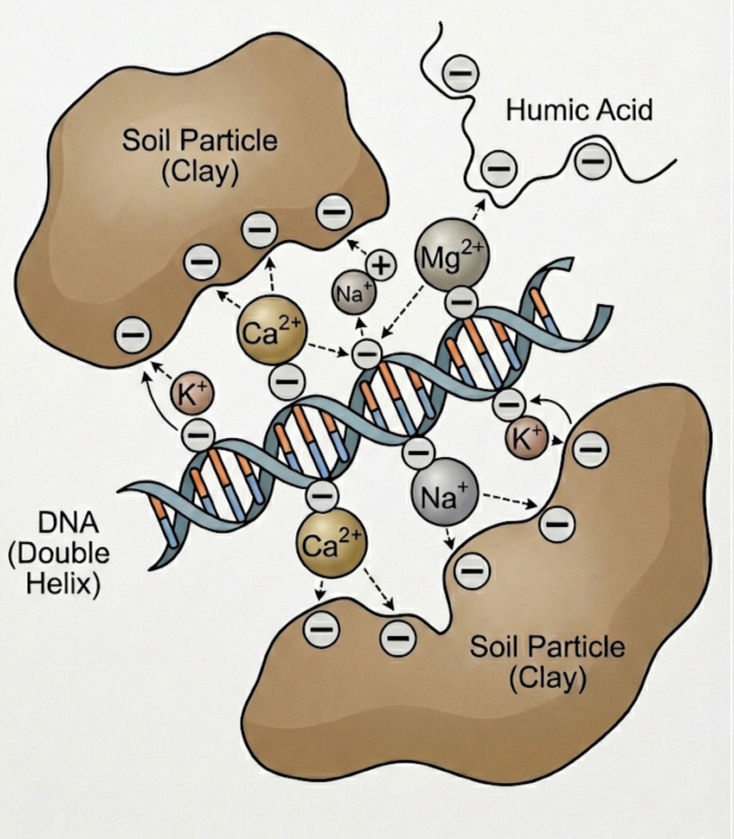
Clays and organic matter are negatively charged and have a high amount of cation exchange capacity -- meaning they can bind positively charged cations. DNA is negatively charged and therefore it will not bind to clays or organic matter as they are both negatively charged. In order for the DNA to be protected, it needs the cations to to first bind to the organic matter and clays, creating a positive binding site for the DNA. This binding mechanism is called a cation bridge.
In summary soils with low organic content and clays, such as sandy soils, will have a low amount of relic DNA while soils with high organic content and clays (high CEC) and have a high amount of cations will bind relic DNA and protect it from degradation.
Relic DNA in Compost, Teas, and Extracts
Compost
Unfortunately, there are currently no studies which address the abundance of relic DNA in compost or vermicompost. However, we can take the lessons learned from soils and try to extrapolate relic DNA in compost. Compost and probably even more so vermicompost have an extremely high amount of organic matter. These molecules can bind tightly to DNA with cation bridges as previously discussed above. However, (vermi)compost is also extremely biologically active and any dead cells would face a very quick turn over and be consumed. So which one of these factors wins out? No one knows yet. In my mind DNA sequencing is likely going to give you an accurate picture of who's there but might be overestimating the amount of organisms present if DNA from dead cells is protected.
Compost Teas and Extracts
Similar to compost there is no studies on relic DNA in compost teas and extracts. However, in the image below you can see that water can have a high proportion of relic DNA.

Teas
Because of the added nutrients in teas, we always see growth or blooms of organisms. In my mind, it's doubtful that relic DNA is an issue because we typically do not see much of the original organisms from the compost. These growing populations of organisms might be consuming dead cells or loose DNA floating around as well.
Extracts
Extracts are a bit trickier because in most cases the extract microbial community reflects the compost community. It is possible that some of these organisms die in the extraction process and it might not be reflected in the DNA sequencing data. For the most part I don't think this is an issue in fresh extract. Additionally, I think the quantitative aspect of Aggrego Data's bacterial sequencing would start to show a decline of organisms if the community began to die off.
Relic DNA Removal
Despite the potential for biases due to relic DNA in almost every biological system, it is not commonly removed before analysis. While Aggrego Data does not currently remove relic DNA, we hope to incorporate into it's removal our DNA extraction methodology in the coming year. I think relic DNA removal is an important step to include for DNA sequencing based methodologies to gain traction in the regulatory environment. It is a potential glaring problem if we want to quantify the total viable organisms, beneficial or pathogenic, in a sample.
Removal techniques
The most common way of removing DNA from dead cells before DNA sequencing is to add Propidium Monoazide (PMA) to the sample before extraction. This molecule will ideally bind to all DNA not protected by a cellular membrane and when exposed to light make the DNA unable to amplified. However, achieving light penetration throughout a soil or compost slurry can be challenging, and it likely does not remove all of the relic DNA. However, a recent 2024 paper demonstrated Benzoase, an enzyme which degrades DNA, has a high efficacy of relic DNA removal and does not require light exposure.
That wraps up this post. If you have any questions or comments about relic DNA or microbial activity please comment below.




Comments